Have you ever searched for your protein in UniProt and found too many results? There's a few strategies that can help narrow down to the right result.
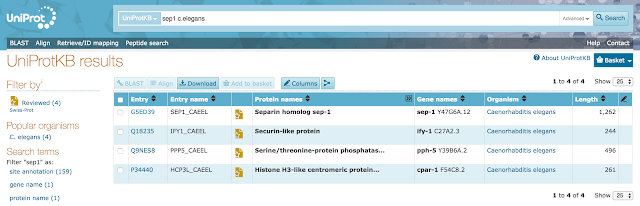
For example, let's say you're searching for proteins belonging to gene SEP1 from the species C. elegans. Let's try searching free-text for 'sep1 c.elegans'. The results bring back 4 proteins and the C. elegans protein for the gene SEP1 is right on top (as of release 2017_07).
You'll notice that the results encompass genes other than SEP1. This is because entering SEP1 as a free-text search will bring back all entries that mention SEP1 anywhere in the text, including unexpected hits for protein entries that are not SEP1 but mention SEP1 as an interactor for example. Nevertheless, your protein of interest appears at the top of the results set.
However when you're searching for something that is likely to bring back a lot more results (like 'Kinase'), you might not be as lucky with free-text searching.
One way of narrowing down your results set is by using the Filters on the UniProtKB results page. The UniProtKB entry page provides a Reviewed/ Unreviewed status filter, Organism filter and a special Search terms filter. You can use the Popular organisms filter to quickly select your desired organism if your query has found results in more than one organisms. The 'Search terms' filter lets you select a category for each of your search terms. For example, here we can choose Search terms "sep1" as 'gene name' to indicate what type of term this is. This will ensure that the website search interprets the search terms in the desired way.
gene:sep1 AND organism:"Caenorhabditis elegans [6239]". This means that you can easily combine both approaches, and you can open any query, whether you built it by typing terms or by using filters, with the advanced search to include additional criteria.
Free-text searching
The most common way to begin a search is to type your search terms directly into the main search bar. Because the UniProtKB search algorithm ranks results on the basis of properties like relevance, annotation score and entry status (reviewed or unreviewed), a free-text search will often return the most relevant hits at the top of your results.
For example, let's say you're searching for proteins belonging to gene SEP1 from the species C. elegans. Let's try searching free-text for 'sep1 c.elegans'. The results bring back 4 proteins and the C. elegans protein for the gene SEP1 is right on top (as of release 2017_07).
You'll notice that the results encompass genes other than SEP1. This is because entering SEP1 as a free-text search will bring back all entries that mention SEP1 anywhere in the text, including unexpected hits for protein entries that are not SEP1 but mention SEP1 as an interactor for example. Nevertheless, your protein of interest appears at the top of the results set.
However when you're searching for something that is likely to bring back a lot more results (like 'Kinase'), you might not be as lucky with free-text searching.
Filtering your search results
One way of narrowing down your results set is by using the Filters on the UniProtKB results page. The UniProtKB entry page provides a Reviewed/ Unreviewed status filter, Organism filter and a special Search terms filter. You can use the Popular organisms filter to quickly select your desired organism if your query has found results in more than one organisms. The 'Search terms' filter lets you select a category for each of your search terms. For example, here we can choose Search terms "sep1" as 'gene name' to indicate what type of term this is. This will ensure that the website search interprets the search terms in the desired way.
Auto-completion for free-text searching
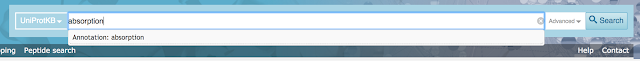
UniProt offers another solution to help you define your search. When you type a search term into the UniProt search box, the site presents an autocompletion suggestion that offers a category to define your term. For example, when you type in 'absorption', you see the autocompletion suggestion 'annotation: absorption'. When you type in 'Ensembl', you see the autocompletion suggestion 'Database:Ensembl' amongst other databases that match your term.
If the suggestion matches the type of category you had in mind, selecting it and then launching the search will help find better matches.
Advanced search for better targeted results
The most powerful way of searching UniProtKB is to use the Advanced search option. It allows you to search for entries by restricting your search to specific categories of data. To bring up the advanced search query builder just click on the 'Advanced' link in the search bar.
Now you can select the categories from a dropdown (the default is 'All' categories) and then specify a related search term. You can enter any number of fields like this and also change the boolean relationships between them (AND, OR, NOT). For example, you could select 'Gene name' and enter SEP1 then in the next row select 'Organism' and enter Caenorhabditis elegans. You will see that an autocompletion functionality brings up a number of suggestions matching your term. Select “Caenorhabditis elegans [6239]”, where 6239 is the taxonomy identifier for Caenorhabditis elegans. Submitting this will return just your one specific search result (the same as when using filters as described above), as opposed to the 4 in the free text search.
The advanced search category dropdown provides a huge number of options in a nested tree structure. For example, if you select 'Function' you will see that there are a number of sub-topics available within it such as 'Active site'. We recommend clicking on the 'help' link in the advanced search widget which takes you to a page with the entire tree structure listed out. This will help you find your topic of choice within the advanced search category dropdown. Note that the search topics (sub-topics) in the advanced search match those in the UniProtKB entry, so it is recommended to familiarize yourself with the entry content to better exploit the search capabilities.
You will note that both approaches – using filters and advanced search – lead you to the same query and URL, namelygene:sep1 AND organism:"Caenorhabditis elegans [6239]". This means that you can easily combine both approaches, and you can open any query, whether you built it by typing terms or by using filters, with the advanced search to include additional criteria.
Using category selections to find a set of proteins
For most search categories, entering text in the term box is optional. If you select 'Active site' and choose to leave the term box blank, you will simply get back all proteins with an 'Active site' associated with them. Thus if you wanted to find all human proteins annotated to be related to a disease, go into the advanced search and select the category 'Organism' and enter human, then select 'Pathology and Biotech' -> 'Disease' and leave the term empty. Hit enter and you will find the results set of all human proteins associated with a disease.
To quickly confirm this, you can click on the 'Columns' button and add the 'Involvement in disease' column to your results page. You can now see all the information about diseases within your results table.






